 |
 |
 |
 |
| Co-PI: Shuyu Liu SLiu@ag.tamu.edu |
Co-PI: Amir Ibrahim, aibrahim@tamu.edu |
Collaborator: Jackie Rudd JCRudd@ag.tamu.edu |
Ph. D. student: Smit Dhakal |
Smit Dhakal is being co-advised by Shuyu Liu and Amir Ibrahim for this project. A major QTL was identified based on a set of 217 F6:7 RILs derived from the cross of CO 960293-2/TAM 111 that were tested in eight environments across Texas, Kansas, Colorado, and Idaho. The genotyping was conducted using 90K array SNP and a set of 5581 SNP were mapped. The major QTL was mapped onto chromosome 2BS that is associated with yield, thousand kernel weight and harvest index with the favorable allele from TAM 111. Released by Texas A&M AgriLife Research in 2003, TAM 111 has been among the most widely grown HRWW cultivars in the Great Plains since 2010. TAM 111 and its derivatives have been used in many wheat breeding programs.
The TX team is focus on a QTL on chromosome arm 2BS associated with yield, thousand kernel weight, and harvest index with favorable allele from TAM 111 (Qgy.tamu.2BS). The QTL was identified in a population (217 F6:7 RILs) derived from the cross of CO 960293-2/TAM 111 evaluated in eight environments across Texas, Kansas, Colorado, and Idaho. The genotyping was conducted using 90K array SNP. The QTL is located at approximately 65.5 Mb in the CS v1 reference and flanking KASP markers were designed (8 KASP in the region). The screening to identify heterogeneous F5 inbred family to develop the larger segregating population is in progress.
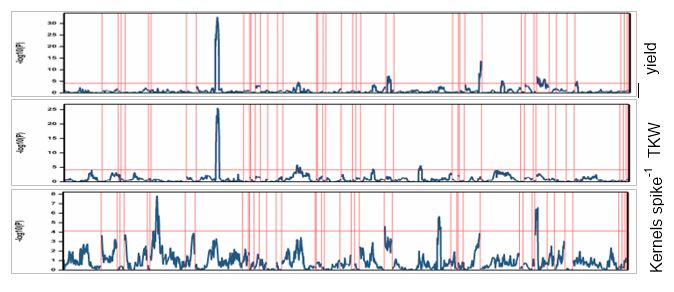
Progress: We have designed 8 KASP SNPs in the 2BS QTL region and 6 KASP SNPs in the 2AL region. These KASP SNPs are being used to confirm the recombinants in the RILs and Screen the F5 lines. A set of 100 F5 lines are being screened by these KASP SNPs.
The QTL on 7AL for kernels spike-1 at 670-680 Mb CSV1 was significant in 3 out of 5 environments. The highest R2 is 7.3% and its additive effect is to increase one seed.
