 |
 |
 |
|
Co-PI: Mark E. Sorrells |
Alvina Gul |
Ph.D. Student: Ella Taagen |
The Cornell Small Grains research program is using the SynOpDH (200) and SynOpRIL (2000) mapping populations to fine-map three QTL for grain weight and grain morphology. The primary targets are a QTL on 5AL for thousand kernel weight (TKW) and grain width at 66 cM, and a QTL on 5BL for grain length at 40.5 cM. In the past two years these QTL have been validated in five different local environments. A secondary target is a seed size QTL positioned more distally on chromosome arm 5BL at ~75 cM, referred to as 5BL2. The donor of the positive allele for the 5AL QTL is Opata whereas the positive allele donor for both 5BL QTL is W7984. The 5AL QTL may be homoeologous to one of the 5BL QTL.
Heterogeneous inbred families (HIFs) were developed from RILs with heterozygous QTL peak flanking markers and self-pollinated for three generations to reduce genetic variability. Phenotypes and genotypes from a 2017 greenhouse planting validated previous results. During the summer of 2018 tissue was harvested from 2,600 HIF plants and fifteen new KASP primers were developed and mapped on the DH population delimiting our three QTL regions of interest. Field-based phenotypes from the 2018 growing season will be recorded for TKW, seed length, width, area and perimeter using a WinSEEDLE scanner. Ella will collect genotypic and phenotypic data from both DH and HIFs and perform analysis using R/QTL this fall in preparation for planting spring head rows. In addition, we are continuing to backcross the positive alleles from the DH population as donors to recurrent parents Opata, Tom and Glenn, and CIMMYT high-biomass lines and high-yielding lines.
| The Sorrells lab has frequently worked with the SynOp mapping population. Previous studies indicate that the 5BL1 QTL has been mapped in four different populations in a total of 18 environments. The studies include an association panel of eastern soft wheat lines over two years in OH and NY (Breseghello & Sorrells 2006), a biparental W7984 x Opata population grown in CA and NY, (Breseghello & Sorrells 2007), a biparental SynOpDH population grown in two GH environments and a field environment (Williams & Sorrells 2014), a biparental cross between two MN lines grown at three locations in MN (Tsilo et al. 2010), and a biparental Renan x Recital grown in six locations in France (Groos et al. 2003). The 5BL2 QTL was mapped in three different populations grown in a total of 15 environments. The 5AL QTL was mapped in three different populations grown in a total of 12 environments. | 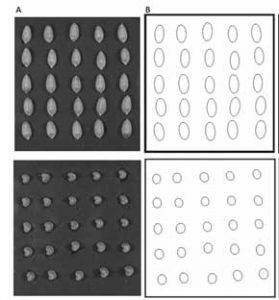
Figure: Image processing via ImageJ: (A) original photograph, (B) transformation to fitted ellipses. |
References
- Breseghello, F. and Sorrells, M.E., 2006. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics, 172(2), pp.1165-1177. DOI: 10.1534/genetics.105.044586
- Breseghello, F. and Sorrells, M.E., 2007. QTL analysis of kernel size and shape in two hexaploid wheat mapping populations. Field crops research, 101(2), pp.172-179. DOI: 10.1016/j.fcr.2006.11.008
- Groos, C., Robert, N., Bervas, E. and Charmet, G., 2003. Genetic analysis of grain protein-content, grain yield and thousand-kernel weight in bread wheat. Theoretical and Applied Genetics, 106(6), pp.1032-1040. DOI: 10.1007/s00122-002-1111-1
- Tsilo, T.J., Hareland, G.A., Simsek, S., Chao, S. and Anderson, J.A., 2010. Genome mapping of kernel characteristics in hard red spring wheat breeding lines. Theoretical and Applied Genetics, 121(4), pp.717-730. DOI: 10.1007/s00122-010-1343-4
